Scientific Research
 The prospect of gaining new insights into the etiology of complex diseases, such as cancers, is rapidly rising with the availability of new measurement technologies, increasing access to large electronic databases and establishment of new cohorts and biobanks. I lead a broad research program in developing and applying quantitative methods for design and analysis of modern large scale biomedical studies with the goals of identifying new risk factors and biomarkers, understanding disease mechanisms, developing models for disease risk prediction and evaluate risk-based strategies for disease prevention. My endeavor in the field has been illustrated through my leadership in design and analysis of a variety of genome-wide association studies (GWAS) that have led to identification new cancer susceptibility SNPs; characterization of genetic architecture and gene-environment interactions; and better understanding of potential for genetic risk stratification for precision cancer prevention. As I had worked at NCI for 16 years, a lot of my subject areas of expertise involves cancers. But many of the concepts, methodologies and tools we have developed are equally applicable to other diseases.
The prospect of gaining new insights into the etiology of complex diseases, such as cancers, is rapidly rising with the availability of new measurement technologies, increasing access to large electronic databases and establishment of new cohorts and biobanks. I lead a broad research program in developing and applying quantitative methods for design and analysis of modern large scale biomedical studies with the goals of identifying new risk factors and biomarkers, understanding disease mechanisms, developing models for disease risk prediction and evaluate risk-based strategies for disease prevention. My endeavor in the field has been illustrated through my leadership in design and analysis of a variety of genome-wide association studies (GWAS) that have led to identification new cancer susceptibility SNPs; characterization of genetic architecture and gene-environment interactions; and better understanding of potential for genetic risk stratification for precision cancer prevention. As I had worked at NCI for 16 years, a lot of my subject areas of expertise involves cancers. But many of the concepts, methodologies and tools we have developed are equally applicable to other diseases.Statistical Genetics
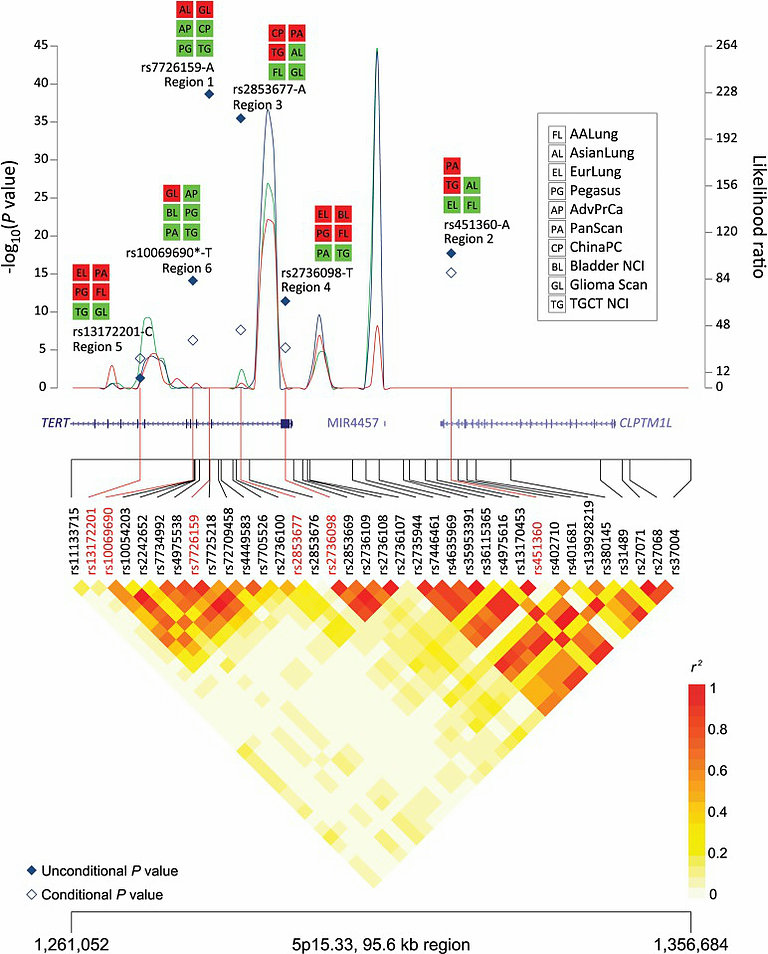 My interest in statistical genetics began when I started my post-doctoral Fellowship at NCI in 1999. I got introduced to basic concepts of genetics while developed methods for estimating risk (penetrance) associated with rare high-penetrant mutations, such as those in the BRCA1/2 genes, using data from the kin-cohort design pioneered by my mentor late, Dr. Sholom Wacholder. Subsequently, I developed an interest in studying gene-environment interactions, specially in the context of epidemiologic case-control design where there is considerable potential for ganing efficiency by novel analytic strategies. I began collaborating in genome-wide association studies from its early days (2006-7) and turned my attention towards a rich set of methodological issues arising from these high throughout agnostic studies. My collaborators and I developed powerful methods for genetic association testing within biologic pathways, meta-analysis accross heterogeneous phenoypes and large-scale exploration of gene-gene and gene-envionment interactions. We also conducted foundational studies establishing mathematical framework to understand potential yield for future GWAS as a function of sample size and genetic architecture of the underlying traits. Applications of these and other methods have led to new insights to genetic basis of cancers as well as a number of other complex traits. We are currently developing methods for integrating various types of functional and annotation information for analysis and interpretation of future GWAS of common and rare variants.
My interest in statistical genetics began when I started my post-doctoral Fellowship at NCI in 1999. I got introduced to basic concepts of genetics while developed methods for estimating risk (penetrance) associated with rare high-penetrant mutations, such as those in the BRCA1/2 genes, using data from the kin-cohort design pioneered by my mentor late, Dr. Sholom Wacholder. Subsequently, I developed an interest in studying gene-environment interactions, specially in the context of epidemiologic case-control design where there is considerable potential for ganing efficiency by novel analytic strategies. I began collaborating in genome-wide association studies from its early days (2006-7) and turned my attention towards a rich set of methodological issues arising from these high throughout agnostic studies. My collaborators and I developed powerful methods for genetic association testing within biologic pathways, meta-analysis accross heterogeneous phenoypes and large-scale exploration of gene-gene and gene-envionment interactions. We also conducted foundational studies establishing mathematical framework to understand potential yield for future GWAS as a function of sample size and genetic architecture of the underlying traits. Applications of these and other methods have led to new insights to genetic basis of cancers as well as a number of other complex traits. We are currently developing methods for integrating various types of functional and annotation information for analysis and interpretation of future GWAS of common and rare variants.Disease risk prediction
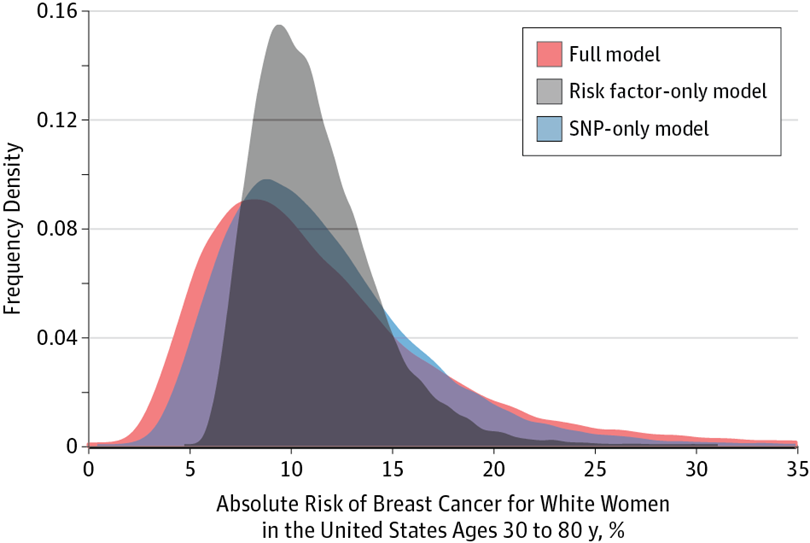 As we develop better understanding of disease risk factors, including genetic susceptibility, biomarkers, environmental and life-style factors, it is critical that such knowledge is translated for developing more effective strategies for disease prevention. A critical step towards this goal is to develop tools that can be used to predict future risk of diseases in healthy individuals and then tailor Disease risk prediction strategies for primary and secondary disease preventions based on individualized risks. Over several years, we have been developing methods for building, validating and assessing risk prediction models that can incorporate both genetic and non-genetic risk factors of diseases. We are studying powerful yet simple methods for constructing predictive polygenic risk score (PRS) using data from GWAS. Most recently, we have developed and distributed a software package, Individualized Coherent Absolute Risk Estimator (iCARE), for building models to predict absolute risks of diseases by synthesizing information from different data sources. We have used this methodology to develop specific models for risk prediction of breast cancer and have assessed clinical utility of such models.
As we develop better understanding of disease risk factors, including genetic susceptibility, biomarkers, environmental and life-style factors, it is critical that such knowledge is translated for developing more effective strategies for disease prevention. A critical step towards this goal is to develop tools that can be used to predict future risk of diseases in healthy individuals and then tailor Disease risk prediction strategies for primary and secondary disease preventions based on individualized risks. Over several years, we have been developing methods for building, validating and assessing risk prediction models that can incorporate both genetic and non-genetic risk factors of diseases. We are studying powerful yet simple methods for constructing predictive polygenic risk score (PRS) using data from GWAS. Most recently, we have developed and distributed a software package, Individualized Coherent Absolute Risk Estimator (iCARE), for building models to predict absolute risks of diseases by synthesizing information from different data sources. We have used this methodology to develop specific models for risk prediction of breast cancer and have assessed clinical utility of such models.General Statistical Methodology
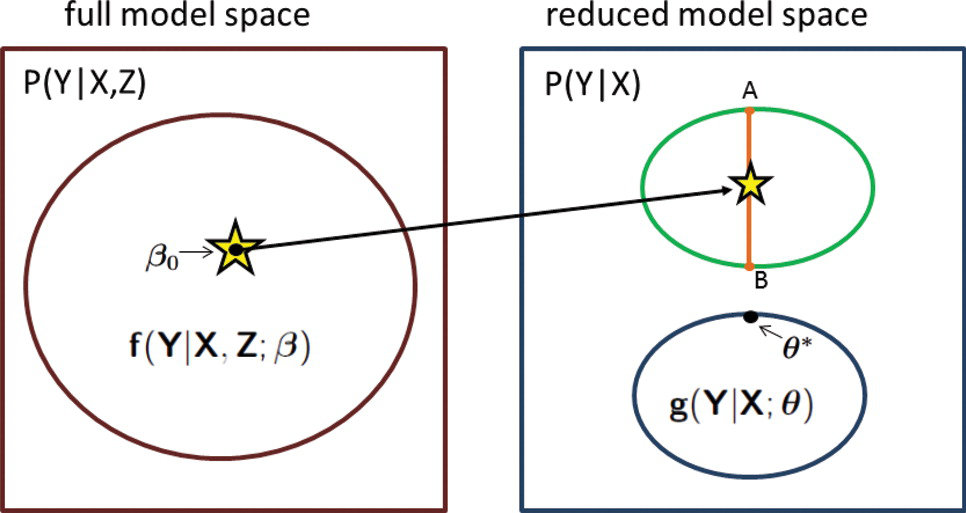 Although a lot of my methodologic research is driven by specific applications, I do maintain an active interest in pursuing more foundational issues faced in statistics in the era of modern data science. Starting from my PhD dissertation research under supervision of late Dr. Norman Breslow, I have maintained a long term interest in studying methods for developing models using studies that employ complex efficient study designs. Examples include the widely used case-control studies that sample subjects based on their disease status and two-phase studies that collect detailed information of potential disease risk factors only on a sub-set of sample of the main study. Most recently, my group has been developing very general methodology for building models using only summary-level information, such as parameters of various sub-models, from multiple heterogeneous data sources. This framework will allow integration of information across different data sources in a statistically and computationally efficient manner without the risks that comes with sharing of individual level data
Although a lot of my methodologic research is driven by specific applications, I do maintain an active interest in pursuing more foundational issues faced in statistics in the era of modern data science. Starting from my PhD dissertation research under supervision of late Dr. Norman Breslow, I have maintained a long term interest in studying methods for developing models using studies that employ complex efficient study designs. Examples include the widely used case-control studies that sample subjects based on their disease status and two-phase studies that collect detailed information of potential disease risk factors only on a sub-set of sample of the main study. Most recently, my group has been developing very general methodology for building models using only summary-level information, such as parameters of various sub-models, from multiple heterogeneous data sources. This framework will allow integration of information across different data sources in a statistically and computationally efficient manner without the risks that comes with sharing of individual level data
